Yeast (Saccharomyces Cerevisiae): Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
'''''Saccharomyces cerevisiae''''' ('''brewer's yeast''' or '''baker's yeast''') is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like ''[[Escherichia coli]]'' as the model bacterium. ''S. cerevisiae'' cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding.<ref>{{cite book|last=Feldmann|first=Horst|title=Yeast. Molecular and Cell bio|date=2010|publisher=Wiley-Blackwell|isbn=978-3527326099}}</ref> | '''''Saccharomyces cerevisiae''''' ('''brewer's yeast''' or '''baker's yeast''') is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like ''[[Escherichia coli]]'' as the model bacterium. ''S. cerevisiae'' cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding.<ref>{{cite book|last=Feldmann|first=Horst|title=Yeast. Molecular and Cell bio|date=2010|publisher=Wiley-Blackwell|isbn=978-3527326099}}</ref> | ||
==Model Organism== | |||
[[File:S cerevisiae under DIC microscopy.jpg|thumb|''S. cerevisiae'', [[Differential interference contrast microscopy|differential interference contrast]] image]] | [[File:S cerevisiae under DIC microscopy.jpg|thumb|''S. cerevisiae'', [[Differential interference contrast microscopy|differential interference contrast]] image]] | ||
[[Image:20100911 232323 Yeast Live.jpg|thumb|''Saccharomyces cerevisiae''<br />Numbered ticks are 11 micrometers apart.]] | [[Image:20100911 232323 Yeast Live.jpg|thumb|''Saccharomyces cerevisiae''<br />Numbered ticks are 11 micrometers apart.]] | ||
| Line 13: | Line 13: | ||
* ''S. cerevisiae'' research is a strong economic driver, at least initially, as a result of its established use in industry. | * ''S. cerevisiae'' research is a strong economic driver, at least initially, as a result of its established use in industry. | ||
==In the Study of Aging== | |||
For more than five decades ''S. cerevisiae'' has been studied as a model organism to better understand aging and has contributed to the identification of more mammalian genes affecting aging than any other model organism.{{pmid|22768836}} Some of the topics studied using yeast are calorie restriction, as well as in genes and cellular pathways involved in senescence. The two most common methods of measuring aging in yeast are Replicative Life Span (RLS), which measures the number of times a cell divides, and Chronological Life Span (CLS), which measures how long a cell can survive in a non-dividing stasis state.{{pmid|22768836}} Limiting the amount of glucose or amino acids in the growth medium has been shown to increase RLS and CLS in yeast as well as other organisms.{{pmid|17530929}} At first, this was thought to increase RLS by up-regulating the sir2 enzyme, however it was later discovered that this effect is independent of sir2. Over-expression of the genes sir2 and fob1 has been shown to increase RLS by preventing the accumulation of extrachromosomal rDNA circles, which are thought to be one of the causes of senescence in yeast.{{pmid|17530929}} The effects of dietary restriction may be the result of a decreased signaling in the TOR cellular pathway.{{pmid|22768836}} This pathway modulates the cell's response to nutrients, and mutations that decrease TOR activity were found to increase CLS and RLS.{{pmid|22768836}}{{pmid|17530929}} This has also been shown to be the case in other animals.{{pmid|22768836}}{{pmid|17530929}} A yeast mutant lacking the genes Sch9 and Ras2 has recently been shown to have a tenfold increase in chronological lifespan under conditions of calorie restriction and is the largest increase achieved in any organism.{{pmid|18225956}} | For more than five decades ''S. cerevisiae'' has been studied as a model organism to better understand aging and has contributed to the identification of more mammalian genes affecting aging than any other model organism.{{pmid|22768836}} Some of the topics studied using yeast are [[Caloric Restriction|calorie restriction]], as well as in genes and cellular pathways involved in [[Senecent Cells|senescence]]. The two most common methods of measuring aging in yeast are Replicative Life Span (RLS), which measures the number of times a cell divides, and Chronological Life Span (CLS), which measures how long a cell can survive in a non-dividing stasis state.{{pmid|22768836}} Limiting the amount of glucose or amino acids in the growth medium has been shown to increase RLS and CLS in yeast as well as other organisms.{{pmid|17530929}} At first, this was thought to increase RLS by up-regulating the [[SIRT2|sir2]] enzyme, however it was later discovered that this effect is independent of sir2. Over-expression of the genes sir2 and fob1 has been shown to increase RLS by preventing the accumulation of extrachromosomal rDNA circles, which are thought to be one of the causes of senescence in yeast.{{pmid|17530929}} The effects of dietary restriction may be the result of a decreased signaling in the TOR cellular pathway.{{pmid|22768836}} This pathway modulates the cell's response to nutrients, and mutations that decrease TOR activity were found to increase CLS and RLS.{{pmid|22768836}}{{pmid|17530929}} This has also been shown to be the case in other animals.{{pmid|22768836}}{{pmid|17530929}} A yeast mutant lacking the genes Sch9 and Ras2 has recently been shown to have a tenfold increase in chronological lifespan under conditions of calorie restriction and is the largest increase achieved in any organism.{{pmid|18225956}} | ||
Mother cells give rise to progeny buds by mitotic divisions, but undergo replicative aging over successive generations and ultimately die. However, when a mother cell undergoes meiosis and gametogenesis, lifespan is reset.{{pmid|21700873}} The replicative potential of gametes (spores) formed by aged cells is the same as gametes formed by young cells, indicating that age-associated damage is removed by meiosis from aged mother cells. This observation suggests that during meiosis removal of age-associated damages leads to rejuvenation. However, the nature of these damages remains to be established. | Mother cells give rise to progeny buds by mitotic divisions, but undergo replicative aging over successive generations and ultimately die. However, when a mother cell undergoes meiosis and gametogenesis, lifespan is reset.{{pmid|21700873}} The replicative potential of gametes (spores) formed by aged cells is the same as gametes formed by young cells, indicating that age-associated damage is removed by meiosis from aged mother cells. This observation suggests that during meiosis removal of age-associated damages leads to rejuvenation. However, the nature of these damages remains to be established. | ||
Revision as of 03:00, 8 December 2023
Saccharomyces cerevisiae (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like Escherichia coli as the model bacterium. S. cerevisiae cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding.[1]
Model Organism
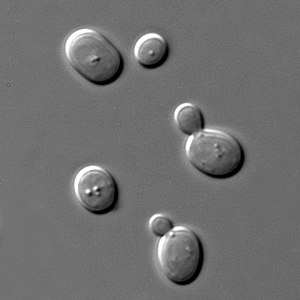
When researchers look for an organism to use in their studies, they look for several traits. Among these are size, generation time, accessibility, manipulation, genetics, conservation of mechanisms, and potential economic benefit. The yeast species S. pombe and S. cerevisiae are both well studied; these two species diverged approximately 600 to 300 million years ago, and are significant tools in the study of DNA damage and repair mechanisms.[2]
S. cerevisiae has developed as a model organism because it scores favorably on a number of these criteria.
- As a single-cell organism, S. cerevisiae is small with a short generation time (doubling time 1.25–2 hours[3] at 30 °C or 86 °F) and can be easily cultured. These are all positive characteristics in that they allow for the swift production and maintenance of multiple specimen lines at low cost.
- S. cerevisiae divides with meiosis, allowing it to be a candidate for sexual genetics research.
- S. cerevisiae can be transformed allowing for either the addition of new genes or deletion through homologous recombination. Furthermore, the ability to grow S. cerevisiae as a haploid simplifies the creation of gene knockout strains.
- As a eukaryote, S. cerevisiae shares the complex internal cell structure of plants and animals without the high percentage of non-coding DNA that can confound research in higher eukaryotes.
- S. cerevisiae research is a strong economic driver, at least initially, as a result of its established use in industry.
In the Study of Aging
For more than five decades S. cerevisiae has been studied as a model organism to better understand aging and has contributed to the identification of more mammalian genes affecting aging than any other model organism.[4] Some of the topics studied using yeast are calorie restriction, as well as in genes and cellular pathways involved in senescence. The two most common methods of measuring aging in yeast are Replicative Life Span (RLS), which measures the number of times a cell divides, and Chronological Life Span (CLS), which measures how long a cell can survive in a non-dividing stasis state.[4] Limiting the amount of glucose or amino acids in the growth medium has been shown to increase RLS and CLS in yeast as well as other organisms.[5] At first, this was thought to increase RLS by up-regulating the sir2 enzyme, however it was later discovered that this effect is independent of sir2. Over-expression of the genes sir2 and fob1 has been shown to increase RLS by preventing the accumulation of extrachromosomal rDNA circles, which are thought to be one of the causes of senescence in yeast.[5] The effects of dietary restriction may be the result of a decreased signaling in the TOR cellular pathway.[4] This pathway modulates the cell's response to nutrients, and mutations that decrease TOR activity were found to increase CLS and RLS.[4][5] This has also been shown to be the case in other animals.[4][5] A yeast mutant lacking the genes Sch9 and Ras2 has recently been shown to have a tenfold increase in chronological lifespan under conditions of calorie restriction and is the largest increase achieved in any organism.[6]
Mother cells give rise to progeny buds by mitotic divisions, but undergo replicative aging over successive generations and ultimately die. However, when a mother cell undergoes meiosis and gametogenesis, lifespan is reset.[7] The replicative potential of gametes (spores) formed by aged cells is the same as gametes formed by young cells, indicating that age-associated damage is removed by meiosis from aged mother cells. This observation suggests that during meiosis removal of age-associated damages leads to rejuvenation. However, the nature of these damages remains to be established.
During starvation of non-replicating S. cerevisiae cells, reactive oxygen species increase leading to the accumulation of DNA damages such as apurinic/apyrimidinic sites and double-strand breaks.[8] Also in non-replicating cells the ability to repair endogenous double-strand breaks declines during chronological aging.[9]
See Also
- Model Organism
- Wikipedia - Saccharomyces cerevisiae
References
- ↑ Feldmann; "Yeast. Molecular and Cell bio" , ISBN: 978-3527326099
- ↑ Nickoloff et al.; "DNA Damage and Repair" , ISBN: 978-1-59259-095-7
- ↑ "Yeasts in Food: Beneficial and Detrimental aspects" , pp. 322 , ISBN: 978-3-86022-961-3
- ↑ Jump up to: 4.0 4.1 4.2 4.3 4.4 Longo VD et al.: Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 2012. (PMID 22768836) [PubMed] [DOI] [Full text] Abstract
- ↑ Jump up to: 5.0 5.1 5.2 5.3 Kaeberlein M et al.: Recent developments in yeast aging. PLoS Genet 2007. (PMID 17530929) [PubMed] [DOI] [Full text] Abstract
- ↑ Wei M et al.: Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet 2008. (PMID 18225956) [PubMed] [DOI] [Full text] Abstract
- ↑ Unal E et al.: Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science 2011. (PMID 21700873) [PubMed] [DOI] [Full text] Abstract
- ↑ Steinboeck F et al.: The relevance of oxidative stress and cytotoxic DNA lesions for spontaneous mutagenesis in non-replicating yeast cells. Mutat Res 2010. (PMID 20223252) [PubMed] [DOI] Abstract
- ↑ Pongpanich M et al.: Pathologic Replication-Independent Endogenous DNA Double-Strand Breaks Repair Defect in Chronological Aging Yeast. Front Genet 2018. (PMID 30410502) [PubMed] [DOI] [Full text] Abstract

