Hallmarks of Aging: Difference between revisions
No edit summary |
|||
| Line 303: | Line 303: | ||
==References== | ==References== | ||
<references /> | <references /> | ||
[[Category:Aging]] | [[Category:Hallmarks of Aging|!Hallmarks_of_Aging]] | ||
Revision as of 10:12, 6 January 2024
Aging is characterized by a progressive loss of physiological integrity, leading to impaired function and increased vulnerability to death. The hallmarks of aging are the types of biochemical changes that occur in all organisms that experience biological aging and lead to a progressive loss of physiological integrity, impaired function and, eventually, death. They were first listed in a landmark paper in 2013[1] to conceptualize the essence of biological aging and its underlying mechanisms.
Criteria
Each hallmark was chosen to try to fulfill the following criteria:[1]
- manifests during normal aging;
- experimentally increasing it accelerates aging;
- experimentally amending it slows the normal aging process and increases healthy lifespan.
These conditions are met to different extents by each of these hallmarks. The last criterion is not present in many of the hallmarks, as science has not yet found feasible ways to amend these problems in living organisms.
Overview
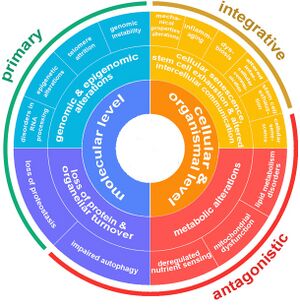
Aging is a complex process characterized by a gradual decline in physiological function. The hallmarks of aging are classified into three categories, each describing different aspects of the aging process:
- Primary Hallmarks: These are considered the main causes of cellular damage leading to aging. They are the initiating factors that, over time, drive the functional decline seen in aging cells and tissues.
- Antagonistic Hallmarks: These hallmarks are the response to the damage caused by the primary hallmarks. Initially, they may be compensatory or protective, but when chronic or excessive, they become deleterious, contributing to the aging process.
- Integrative Hallmarks: These hallmarks are the culprits of the phenotype of aging. They result from a combination of the primary and antagonistic hallmarks and are ultimately responsible for the functional decline in tissues and organs seen in aging.
The Nine Hallmarks of Aging (2013)
The nine hallmarks of aging were originally conceptualized by López-Otín and colleagues in 2013[1]. Since that it has served as a foundational paradigm for aging research for a decade until it has been revised in 2022. The original 9 landmarks were defined as follows:
| # | Hallmark | |
|---|---|---|
| 1 | Genomic Instability | Primary Hallmarks
(causes damage) |
| 2 | Telomere Attrition | |
| 3 | Epigenetic Alterations | |
| 4 | Loss of Proteostasis | |
| 5 | Deregulated Nutrient Sensing | Antagonistic Hallmarks
(responses to damage) |
| 6 | Mitochondrial Dysfunction | |
| 7 | Cellular Senescence | |
| 8 | Stem Cell Exhaustion | Integrative Hallmarks
(culprits of the phenotype) |
| 9 | Altered Intercellular Communication |
Five New Hallmarks of Aging (2022)
While these nine hallmarks have significantly advanced our understanding of aging and its relation to age-related diseases, recent critiques and evolving scientific evidence have prompted the scientific community to reconsider and expand this framework.[4] To address this, a symposium titled “New Hallmarks of Ageing” was held in Copenhagen on March 2022, where leading experts gathered to discuss potential additions and recontextualizations of these aging hallmarks. The symposium highlighted the critical need for an expanded, inclusive paradigm that encompasses newly identified processes contributing to aging. The discussions suggested the integration of five additional hallmarks:
- compromised autophagy
- dysregulation in RNA splicing
- inflammation
- loss of cytoskeleton integrity
- disturbance of the microbiome (dysbiosis)
These potential new hallmarks, along with the original nine, underscore a more comprehensive understanding of the aging process, acknowledging its multifaceted nature and its profound implications for human health and longevity.[5]
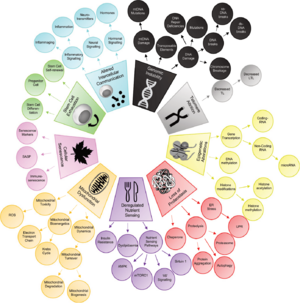
The Twelve Hallmarks of Aging (2023)
The original authors of the nine hallmarks of aging update the set of proposed hallmarks after a decade.[6]
| Level | Hallmark | Description | Proposed Year |
Category | |
|---|---|---|---|---|---|
| Molecular Level |

|
Genomic instability | Accumulation of DNA damage over time leading to cellular dysfunction. | 2013 | Primary Hallmarks (causes damage) |
|
Telomere attrition | Reduction in the length of telomeres leading to cellular aging. | 2013 | ||

|
Epigenetic alterations | Changes in DNA methylation and histone modification affecting gene expression. | 2013 | ||
|
Loss of proteostasis | Disruption in protein folding and stability leading to cell damage. | 2013 | ||

|
Disabled macroautophagy | Impaired cellular maintenance through the consumption of own components. | 2021[7] | Antagonistic Hallmarks (responses to damage) | |
| Cellular & Organismal Level |

|
Deregulated nutrient sensing | Alterations in nutrient sensing pathways affecting metabolism and aging. | 2013 | |
|
Mitochondrial dysfunction | Decrease in mitochondrial efficiency and increase in oxidative stress. | 2013 | ||

|
Cellular senescence | Accumulation of non-dividing, dysfunctional cells secreting harmful factors. | 2013 | ||
|
Stem cell exhaustion | Decline in the regenerative capacity of stem cells affecting tissue repair. | 2013 | Integrative Hallmarks (culprits of the phenotype) | |
|
Dysbiosis
(Microbiome disturbance) |
Changes in gut microbiome affecting health and aging. | |||

|
Chronic inflammation
(Inflammaging) |
Systemic inflammation contributing to aging and related diseases. | 2023[8] | ||

|
Altered intercellular communication | Changes in cellular communication leading to inflammation and tissue dysfunction. | 2013 | ||
Potential Hallmarks of Aging

|
Altered mechanical properties | Changes in cellular and extracellular structure affecting tissue function. | |

|
Splicing dysregulation
(Dysregulation in RNA splicing) |
Impaired RNA construction from DNA, leading to cellular dysfunction. | 2019[9] |
The Hallmarks in Detail
| Hallmark | Background | Manifests during normal aging | Experimentally increasing it accelerates aging | Experimentally amending it slows the normal aging process and increases healthy lifespan. | Associated human diseases |
|---|---|---|---|---|---|
Genomic instability
|
Damange in the DNA are formed mainly through oxidative stress and environmental factors.[10] A number of molecular processes work continuously to repair this damage.[11] | DNA damage accumulates over time[12] | Deficient DNA repair causes premature aging[13] | Increased DNA repair facilitates greater longevity[13] | |
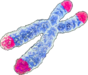
Telomere attrition
|
Telomere attrition refers to the progressive shortening of telomeres, which are protective sequences at the ends of chromosomes. This occurs due to the inability of DNA polymerases to completely replicate the ends of linear DNA, and the absence of telomerase in most somatic cells. Shortened telomeres lead to cellular aging and reduced regenerative capacity, manifesting as replicative senescence or Hayflick limit[14]. Shelterins protect telomeres but may mask damage leading to persistent DNA damage and cellular stress[15]. Dysfunctions in telomere maintenance are linked to various age-related diseases[16]. | Telomere shortening is observed during normal aging in humans and mice[17]. | Excessive telomere attrition due to stress or genetic factors accelerates cellular aging and the onset of age-related pathologies[18]. | Maintaining telomere length through telomerase activation or shelterin integrity can delay aging and extend lifespan, as shown in mouse models and suggested by human epidemiological studies[19][20]. | Telomere shortening is associated with a variety of human diseases, including pulmonary fibrosis, dyskeratosis congenita, and aplastic anemia, often linked to deficiencies in telomerase or shelterin components[16]. |
Epigenetic alterations
|
Histone modifications are a type of epigenetic alteration that play a crucial role in regulating gene expression. Histones are proteins around which DNA is wrapped in eukaryotic cells, forming a structure known as a nucleosome. These modifications occur primarily at the tails of histone proteins and influence how tightly or loosely DNA is wound around the histones, affecting the accessibility of the DNA to various cellular machinery for processes like transcription, replication, and repair. | Chemical changes to histone proteins after they are formed can activate or silence gene expression and regulate the aging process.[21] | Sirtuins influence histone modifications:
|
Disorders in histone modification are linked with various aging-related conditions, implicating altered gene expression and protein function[26]. | |
| DNA methylation shift: DNA methylation is a biochemical process involving the addition of a methyl group to the DNA molecule, specifically to the cytosine or adenine DNA nucleotides. This process is a form of epigenetic modification, which means it can affect gene expression and function without changing the DNA sequence itself. | DNA methylation generally decreases with age in certain human and mouse tissues or cell cultures.[27][28][29][30] The loss of methylation in CD4+ T cells is proportional to age.[28] | No direct evidence yet. | No direct evidence yet. | Progeroid syndromes exhibit DNA methylation patterns similar to normal aging, suggesting a link with aging-related diseases[31][32]. | |
| Chromatin remodeling in the context of epigenetic alterations refers to the dynamic modification of the chromatin architecture to regulate access to genetic information in the DNA. Chromatin, which consists of DNA wrapped around histone proteins, can be altered or remodeled in various ways to either condense and silence gene regions or relax and activate them. This remodeling is a crucial mechanism for controlling gene expression, replication, repair, and other essential cellular processes. | Global canonical histone loss is regarded as a common feature of aging from yeast to humans.[33][34][35] | Flies with loss-of-function mutations in HP1α (a key chromosomal protein) have a shortened lifespan.[26] | Overexpression of HP1α extends longevity in flies and delays the muscular deterioration characteristic of old age.[26] | ||
| Transcriptional alterations refer to changes in the process by which genes are expressed into RNA, which can then be translated into proteins. These alterations can influence which genes are turned on or off, the timing of their activation, the amount of RNA produced, and the stability or processing of that RNA. | Aging leads to more transcriptional noise, meaning more random variations in gene activity[36], and also causes irregular production and processing of many mRNAs, the molecules that carry genetic information from DNA for protein creation[37][38]. Also miRNAs that is associated with the aging process is affected[39][40]. | Expression of several miRNAs increases longevity in Drosophila melanogaster and C. elegans[41][42][43]. | Loss of several miRNAs is associated with increase aging in Drosophila melanogaster and C. elegans[41][42][43]. | ||
Loss of proteostasis
|
Loss of proteostasis refers to the disruption of the body's ability to regulate its proteins effectively, a process in which chaperones play a crucial role. This encompasses the processes of protein synthesis, folding, transport, and degradation. Chaperones, specialized proteins that assist in the proper folding and stabilization of other proteins, are essential in maintaining proteostasis. As we age, or in certain diseases, the balance of these processes can be disturbed, leading to the accumulation of misfolded or damaged proteins, and a decrease in the ability to produce and maintain healthy proteins. The decline in the efficiency or availability of chaperones contributes to this loss of proteostasis, exacerbating the accumulation of dysfunctional proteins and cellular stress. | Aging and some aging-related diseases are linked to impaired protein homeostasis or proteostasis.[44] | Mutant mice that that lack a certain helper chaperone from the heat-shock protein family show accelerated aging.[45] | Transgenic worms and flies overexpressing chaperones are long-lived[46][47] | Chronic expression of unfolded, misfolded or aggregated proteins contributes to the development of some age-related pathologies, such as Alzheimer’s disease, Parkinson’s disease and cataracts[44]. |
Disabled macroautophagy
|
Disabled macroautophagy, often referred as impaired or dysfunctional autophagy, is a condition where the cellular process of autophagy—specifically the macroautophagy pathway—is disrupted or less effective. Autophagy is a critical cellular process for degrading and recycling damaged organelles, misfolded proteins, and other cellular debris. Macroautophagy involves the engulfment of these unwanted materials into vesicles called autophagosomes, which then fuse with lysosomes where the contents are degraded and recycled. When macroautophagy is disabled or impaired, cells accumulate damaged proteins and organelles, leading to cellular dysfunction and contributing to various diseases, particularly those related to aging and neurodegeneration. This loss of a crucial cellular "cleanup" mechanism can result in increased oxidative stress, disrupted cellular homeostasis, and an acceleration of the aging process.
While originally considered under hallmark altered proteostasis, autophagy regulates a number of other hallmarks of ageing such as DNA repair and nutrient sensing/metabolism[48], and hence it was proposed to be categorized as an integrative hallmark. |
Compromised autophagy is observed in numerous ageing conditions including neurodegeneration and immunosenescence[49][50]. | Activation of autophagy can increase mouse lifespan[51], and even improve immune response to vaccination in older humans by overcoming immunosenescence[52]. | ||
Deregulated nutrient sensing
|
|||||
Mitochondrial dysfunction
|
|||||
Cellular senescence
|
|||||
Stem cell exhaustion
|
|||||
| Dysbiosis | |||||
| Chronic inflammation | |||||
Altered intercellular communication
|
History
- 2013 The scientific journal Cell published the article "The Hallmarks of Aging", that was translated to several languages and determined the directions of many studies.[1]
- 2022 It was proposed to expand the list of the nine hallmarks of aging with five more.[5][53]
- 2023 In a paywalled review, the authors of a heavily cited paper on the hallmarks of aging update the set of proposed hallmarks after a decade.[6] A review with overlapping authors merge or link various hallmarks of cancer with those of aging.[54]
Todo
- 2013, The hallmarks of aging [1]
- 2023, Chronic inflammation and the hallmarks of aging [8]
- 2023, Aging Hallmarks and Progression and Age-Related Diseases: A Landscape View of Research Advancement [2]
- 2022, Biological mechanisms of aging predict age-related disease co-occurrence in patients [3]
- https://doi.org/10.1038/s41587-023-02024-y
References
- ↑ 1.0 1.1 1.2 1.3 1.4 López-Otín C et al.: The hallmarks of aging. Cell 2013. (PMID 23746838) [PubMed] [DOI] [Full text] Aging is characterized by a progressive loss of physiological integrity, leading to impaired function and increased vulnerability to death. This deterioration is the primary risk factor for major human pathologies, including cancer, diabetes, cardiovascular disorders, and neurodegenerative diseases. Aging research has experienced an unprecedented advance over recent years, particularly with the discovery that the rate of aging is controlled, at least to some extent, by genetic pathways and biochemical processes conserved in evolution. This Review enumerates nine tentative hallmarks that represent common denominators of aging in different organisms, with special emphasis on mammalian aging. These hallmarks are: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication. A major challenge is to dissect the interconnectedness between the candidate hallmarks and their relative contributions to aging, with the final goal of identifying pharmaceutical targets to improve human health during aging, with minimal side effects.
- ↑ 2.0 2.1 Tenchov R et al.: Aging Hallmarks and Progression and Age-Related Diseases: A Landscape View of Research Advancement. ACS Chem Neurosci 2023. (PMID 38095562) [PubMed] [DOI] Aging is a dynamic, time-dependent process that is characterized by a gradual accumulation of cell damage. Continual functional decline in the intrinsic ability of living organisms to accurately regulate homeostasis leads to increased susceptibility and vulnerability to diseases. Many efforts have been put forth to understand and prevent the effects of aging. Thus, the major cellular and molecular hallmarks of aging have been identified, and their relationships to age-related diseases and malfunctions have been explored. Here, we use data from the CAS Content Collection to analyze the publication landscape of recent aging-related research. We review the advances in knowledge and delineate trends in research advancements on aging factors and attributes across time and geography. We also review the current concepts related to the major aging hallmarks on the molecular, cellular, and organismic level, age-associated diseases, with attention to brain aging and brain health, as well as the major biochemical processes associated with aging. Major age-related diseases have been outlined, and their correlations with the major aging features and attributes are explored. We hope this review will be helpful for apprehending the current knowledge in the field of aging mechanisms and progression, in an effort to further solve the remaining challenges and fulfill its potential.
- ↑ 3.0 3.1 Fraser HC et al.: Biological mechanisms of aging predict age-related disease co-occurrence in patients. Aging Cell 2022. (PMID 35259281) [PubMed] [DOI] [Full text] Genetic, environmental, and pharmacological interventions into the aging process can confer resistance to multiple age-related diseases in laboratory animals, including rhesus monkeys. These findings imply that individual mechanisms of aging might contribute to the co-occurrence of age-related diseases in humans and could be targeted to prevent these conditions simultaneously. To address this question, we text mined 917,645 literature abstracts followed by manual curation and found strong, non-random associations between age-related diseases and aging mechanisms in humans, confirmed by gene set enrichment analysis of GWAS data. Integration of these associations with clinical data from 3.01 million patients showed that age-related diseases associated with each of five aging mechanisms were more likely than chance to be present together in patients. Genetic evidence revealed that innate and adaptive immunity, the intrinsic apoptotic signaling pathway and activity of the ERK1/2 pathway were associated with multiple aging mechanisms and diverse age-related diseases. Mechanisms of aging hence contribute both together and individually to age-related disease co-occurrence in humans and could potentially be targeted accordingly to prevent multimorbidity.
- ↑ Gems D & de Magalhães JP: The hoverfly and the wasp: A critique of the hallmarks of aging as a paradigm. Ageing Res Rev 2021. (PMID 34271186) [PubMed] [DOI] [Full text] With the goal of representing common denominators of aging in different organisms López-Otín et al. in 2013 described nine hallmarks of aging. Since then, this representation has become a major reference point for the biogerontology field. The template for the hallmarks of aging account originated from landmark papers by Hanahan and Weinberg (2000, 2011) defining first six and later ten hallmarks of cancer. Here we assess the strengths and weaknesses of the hallmarks of aging account. As a checklist of diverse major foci of current aging research, it has provided a useful shared overview for biogerontology during a time of transition in the field. It also seems useful in applied biogerontology, to identify interventions (e.g. drugs) that impact multiple symptomatic features of aging. However, while the hallmarks of cancer provide a paradigmatic account of the causes of cancer with profound explanatory power, the hallmarks of aging do not. A worry is that as a non-paradigm the hallmarks of aging have obscured the urgent need to define a genuine paradigm, one that can provide a useful basis for understanding the mechanistic causes of the diverse aging pathologies. We argue that biogerontology must look and move beyond the hallmarks to understand the process of aging.
- ↑ 5.0 5.1 Schmauck-Medina T et al.: New hallmarks of ageing: a 2022 Copenhagen ageing meeting summary. Aging (Albany NY) 2022. (PMID 36040386) [PubMed] [DOI] [Full text] Genomic instability, telomere attrition, epigenetic alterations, mitochondrial dysfunction, loss of proteostasis, deregulated nutrient-sensing, cellular senescence, stem cell exhaustion, and altered intercellular communication were the original nine hallmarks of ageing proposed by López-Otín and colleagues in 2013. The proposal of these hallmarks of ageing has been instrumental in guiding and pushing forward research on the biology of ageing. In the nearly past 10 years, our in-depth exploration on ageing research has enabled us to formulate new hallmarks of ageing which are compromised autophagy, microbiome disturbance, altered mechanical properties, splicing dysregulation, and inflammation, among other emerging ones. Amalgamation of the 'old' and 'new' hallmarks of ageing may provide a more comprehensive explanation of ageing and age-related diseases, shedding light on interventional and therapeutic studies to achieve healthy, happy, and productive lives in the elderly.
- ↑ 6.0 6.1 López-Otín C et al.: Hallmarks of aging: An expanding universe. Cell 2023. (PMID 36599349) [PubMed] [DOI] Aging is driven by hallmarks fulfilling the following three premises: (1) their age-associated manifestation, (2) the acceleration of aging by experimentally accentuating them, and (3) the opportunity to decelerate, stop, or reverse aging by therapeutic interventions on them. We propose the following twelve hallmarks of aging: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, disabled macroautophagy, deregulated nutrient-sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, altered intercellular communication, chronic inflammation, and dysbiosis. These hallmarks are interconnected among each other, as well as to the recently proposed hallmarks of health, which include organizational features of spatial compartmentalization, maintenance of homeostasis, and adequate responses to stress.
- ↑ Kaushik S et al.: Autophagy and the hallmarks of aging. Ageing Res Rev 2021. (PMID 34563704) [PubMed] [DOI] [Full text] Autophagy, an essential cellular process that mediates degradation of proteins and organelles in lysosomes, has been tightly linked to cellular quality control for its role as part of the proteostasis network. The current interest in identifying the cellular and molecular determinants of aging, has highlighted the important contribution of malfunctioning of autophagy with age to the loss of proteostasis that characterizes all old organisms. However, the diversity of cellular functions of the different types of autophagy and the often reciprocal interactions of autophagy with other determinants of aging, is placing autophagy at the center of the aging process. In this work, we summarize evidence for the contribution of autophagy to health- and lifespan and provide examples of the bidirectional interplay between autophagic pathways and several of the so-called hallmarks of aging. This central role of autophagy in aging, and the dependence on autophagy of many geroprotective interventions, has motivated a search for direct modulators of autophagy that could be used to slow aging and extend healthspan. Here, we review some of those ongoing therapeutic efforts and comment on the potential of targeting autophagy in aging.
- ↑ 8.0 8.1 Baechle JJ et al.: Chronic inflammation and the hallmarks of aging. Mol Metab 2023. (PMID 37329949) [PubMed] [DOI] [Full text] BACKGROUND: Recently, the hallmarks of aging were updated to include dysbiosis, disabled macroautophagy, and chronic inflammation. In particular, the low-grade chronic inflammation during aging, without overt infection, is defined as "inflammaging," which is associated with increased morbidity and mortality in the aging population. Emerging evidence suggests a bidirectional and cyclical relationship between chronic inflammation and the development of age-related conditions, such as cardiovascular diseases, neurodegeneration, cancer, and frailty. How the crosstalk between chronic inflammation and other hallmarks of aging underlies biological mechanisms of aging and age-related disease is thus of particular interest to the current geroscience research. SCOPE OF REVIEW: This review integrates the cellular and molecular mechanisms of age-associated chronic inflammation with the other eleven hallmarks of aging. Extra discussion is dedicated to the hallmark of "altered nutrient sensing," given the scope of Molecular Metabolism. The deregulation of hallmark processes during aging disrupts the delicate balance between pro-inflammatory and anti-inflammatory signaling, leading to a persistent inflammatory state. The resultant chronic inflammation, in turn, further aggravates the dysfunction of each hallmark, thereby driving the progression of aging and age-related diseases. MAIN CONCLUSIONS: The crosstalk between chronic inflammation and other hallmarks of aging results in a vicious cycle that exacerbates the decline in cellular functions and promotes aging. Understanding this complex interplay will provide new insights into the mechanisms of aging and the development of potential anti-aging interventions. Given their interconnectedness and ability to accentuate the primary elements of aging, drivers of chronic inflammation may be an ideal target with high translational potential to address the pathological conditions associated with aging.
- ↑ Bhadra M et al.: Alternative splicing in aging and longevity. Hum Genet 2020. (PMID 31834493) [PubMed] [DOI] [Full text] Alternative pre-mRNA splicing increases the complexity of the proteome that can be generated from the available genomic coding sequences. Dysregulation of the splicing process has been implicated in a vast repertoire of diseases. However, splicing has recently been linked to both the aging process itself and pro-longevity interventions. This review focuses on recent research towards defining RNA splicing as a new hallmark of aging. We highlight dysfunctional alternative splicing events that contribute to the aging phenotype across multiple species, along with recent efforts toward deciphering mechanistic roles for RNA splicing in the regulation of aging and longevity. Further, we discuss recent research demonstrating a direct requirement for specific splicing factors in pro-longevity interventions, and specifically how nutrient signaling pathways interface to splicing factor regulation and downstream splicing targets. Finally, we review the emerging potential of using splicing profiles as a predictor of biological age and life expectancy. Understanding the role of RNA splicing components and downstream targets altered in aging may provide opportunities to develop therapeutics and ultimately extend healthy lifespan in humans.
- ↑ De Bont R & van Larebeke N: Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 2004. (PMID 15123782) [PubMed] [DOI] DNA damage plays a major role in mutagenesis, carcinogenesis and ageing. The vast majority of mutations in human tissues are certainly of endogenous origin. A thorough knowledge of the types and prevalence of endogenous DNA damage is thus essential for an understanding of the interactions of endogenous processes with exogenous agents and the influence of damage of endogenous origin on the induction of cancer and other diseases. In particular, this seems important in risk evaluation concerning exogenous agents that also occur endogenously or that, although chemically different from endogenous ones, generate the same DNA adducts. This knowledge may also be crucial to the development of rational chemopreventive strategies. A list of endogenous DNA-damaging agents, processes and DNA adduct levels is presented. For the sake of comparison, DNA adduct levels are expressed in a standardized way, including the number of adducts per 10(6) nt. This list comprises numerous reactive oxygen species and products generated as a consequence (e.g. lipid peroxides), endogenous reactive chemicals (e.g. aldehydes and S-adenosylmethionine), and chemical DNA instability (e.g. depurination). The respective roles of endogenous versus exogenous DNA damage in carcinogenesis are discussed.
- ↑ de Duve C: The onset of selection. Nature 2005. (PMID 15703726) [PubMed] [DOI]
- ↑ Vijg J & Suh Y: Genome instability and aging. Annu Rev Physiol 2013. (PMID 23398157) [PubMed] [DOI] Genome instability has long been implicated as the main causal factor in aging. Somatic cells are continuously exposed to various sources of DNA damage, from reactive oxygen species to UV radiation to environmental mutagens. To cope with the tens of thousands of chemical lesions introduced into the genome of a typical cell each day, a complex network of genome maintenance systems acts to remove damage and restore the correct base pair sequence. Occasionally, however, repair is erroneous, and such errors, as well as the occasional failure to correctly replicate the genome during cell division, are the basis for mutations and epimutations. There is now ample evidence that mutations accumulate in various organs and tissues of higher animals, including humans, mice, and flies. What is not known, however, is whether the frequency of these random changes is sufficient to cause the phenotypic effects generally associated with aging. The exception is cancer, an age-related disease caused by the accumulation of mutations and epimutations. Here, we first review current concepts regarding the relationship between DNA damage, repair, and mutation, as well as the data regarding genome alterations as a function of age. We then describe a model for how randomly induced DNA sequence and epigenomic variants in the somatic genomes of animals can result in functional decline and disease in old age. Finally, we discuss the genetics of genome instability in relation to longevity to address the importance of alterations in the somatic genome as a causal factor in aging and to underscore the opportunities provided by genetic approaches to develop interventions that attenuate genome instability, reduce disease risk, and increase life span.
- ↑ 13.0 13.1 Hoeijmakers JH: DNA damage, aging, and cancer. N Engl J Med 2009. (PMID 19812404) [PubMed] [DOI]
- ↑ Blackburn EH et al.: Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 2006. (PMID 17024208) [PubMed] [DOI]
- ↑ Palm W & de Lange T: How shelterin protects mammalian telomeres. Annu Rev Genet 2008. (PMID 18680434) [PubMed] [DOI] The genomes of prokaryotes and eukaryotic organelles are usually circular as are most plasmids and viral genomes. In contrast, the nuclear genomes of eukaryotes are organized on linear chromosomes, which require mechanisms to protect and replicate DNA ends. Eukaryotes navigate these problems with the advent of telomeres, protective nucleoprotein complexes at the ends of linear chromosomes, and telomerase, the enzyme that maintains the DNA in these structures. Mammalian telomeres contain a specific protein complex, shelterin, that functions to protect chromosome ends from all aspects of the DNA damage response and regulates telomere maintenance by telomerase. Recent experiments, discussed here, have revealed how shelterin represses the ATM and ATR kinase signaling pathways and hides chromosome ends from nonhomologous end joining and homology-directed repair.
- ↑ 16.0 16.1 Armanios M & Blackburn EH: The telomere syndromes. Nat Rev Genet 2012. (PMID 22965356) [PubMed] [DOI] [Full text] There has been mounting evidence of a causal role for telomere dysfunction in a number of degenerative disorders. Their manifestations encompass common disease states such as idiopathic pulmonary fibrosis and bone marrow failure. Although these disorders seem to be clinically diverse, collectively they comprise a single syndrome spectrum defined by the short telomere defect. Here we review the manifestations and unique genetics of telomere syndromes. We also discuss their underlying molecular mechanisms and significance for understanding common age-related disease processes.
- ↑ Blasco MA: Telomere length, stem cells and aging. Nat Chem Biol 2007. (PMID 17876321) [PubMed] [DOI] Telomere shortening occurs concomitant with organismal aging, and it is accelerated in the context of human diseases associated with mutations in telomerase, such as some cases of dyskeratosis congenita, idiopathic pulmonary fibrosis and aplastic anemia. People with these diseases, as well as Terc-deficient mice, show decreased lifespan coincidental with a premature loss of tissue renewal, which suggests that telomerase is rate-limiting for tissue homeostasis and organismal survival. These findings have gained special relevance as they suggest that telomerase activity and telomere length can directly affect the ability of stem cells to regenerate tissues. If this is true, stem cell dysfunction provoked by telomere shortening may be one of the mechanisms responsible for organismal aging in both humans and mice. Here, we will review the current evidence linking telomere shortening to aging and stem cell dysfunction.
- ↑ Fumagalli M et al.: Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 2012. (PMID 22426077) [PubMed] [DOI] [Full text] The DNA-damage response (DDR) arrests cell-cycle progression until damage is removed. DNA-damage-induced cellular senescence is associated with persistent DDR. The molecular bases that distinguish transient from persistent DDR are unknown. Here we show that a large fraction of exogenously induced persistent DDR markers is associated with telomeric DNA in cultured cells and mammalian tissues. In yeast, a chromosomal DNA double-strand break next to a telomeric sequence resists repair and impairs DNA ligase 4 recruitment. In mammalian cells, ectopic localization of telomeric factor TRF2 next to a double-strand break induces persistent DNA damage and DDR. Linear, but not circular, telomeric DNA or scrambled DNA induces a prolonged checkpoint in normal cells. In terminally differentiated tissues of old primates, DDR markers accumulate at telomeres that are not critically short. We propose that linear genomes are not uniformly reparable and that telomeric DNA tracts, if damaged, are irreparable and trigger persistent DDR and cellular senescence.
- ↑ Jaskelioff M et al.: Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 2011. (PMID 21113150) [PubMed] [DOI] [Full text] An ageing world population has fuelled interest in regenerative remedies that may stem declining organ function and maintain fitness. Unanswered is whether elimination of intrinsic instigators driving age-associated degeneration can reverse, as opposed to simply arrest, various afflictions of the aged. Such instigators include progressively damaged genomes. Telomerase-deficient mice have served as a model system to study the adverse cellular and organismal consequences of wide-spread endogenous DNA damage signalling activation in vivo. Telomere loss and uncapping provokes progressive tissue atrophy, stem cell depletion, organ system failure and impaired tissue injury responses. Here, we sought to determine whether entrenched multi-system degeneration in adult mice with severe telomere dysfunction can be halted or possibly reversed by reactivation of endogenous telomerase activity. To this end, we engineered a knock-in allele encoding a 4-hydroxytamoxifen (4-OHT)-inducible telomerase reverse transcriptase-oestrogen receptor (TERT-ER) under transcriptional control of the endogenous TERT promoter. Homozygous TERT-ER mice have short dysfunctional telomeres and sustain increased DNA damage signalling and classical degenerative phenotypes upon successive generational matings and advancing age. Telomerase reactivation in such late generation TERT-ER mice extends telomeres, reduces DNA damage signalling and associated cellular checkpoint responses, allows resumption of proliferation in quiescent cultures, and eliminates degenerative phenotypes across multiple organs including testes, spleens and intestines. Notably, somatic telomerase reactivation reversed neurodegeneration with restoration of proliferating Sox2(+) neural progenitors, Dcx(+) newborn neurons, and Olig2(+) oligodendrocyte populations. Consistent with the integral role of subventricular zone neural progenitors in generation and maintenance of olfactory bulb interneurons, this wave of telomerase-dependent neurogenesis resulted in alleviation of hyposmia and recovery of innate olfactory avoidance responses. Accumulating evidence implicating telomere damage as a driver of age-associated organ decline and disease risk and the marked reversal of systemic degenerative phenotypes in adult mice observed here support the development of regenerative strategies designed to restore telomere integrity.
- ↑ Bernardes de Jesus B et al.: Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med 2012. (PMID 22585399) [PubMed] [DOI] [Full text] A major goal in aging research is to improve health during aging. In the case of mice, genetic manipulations that shorten or lengthen telomeres result, respectively, in decreased or increased longevity. Based on this, we have tested the effects of a telomerase gene therapy in adult (1 year of age) and old (2 years of age) mice. Treatment of 1- and 2-year old mice with an adeno associated virus (AAV) of wide tropism expressing mouse TERT had remarkable beneficial effects on health and fitness, including insulin sensitivity, osteoporosis, neuromuscular coordination and several molecular biomarkers of aging. Importantly, telomerase-treated mice did not develop more cancer than their control littermates, suggesting that the known tumorigenic activity of telomerase is severely decreased when expressed in adult or old organisms using AAV vectors. Finally, telomerase-treated mice, both at 1-year and at 2-year of age, had an increase in median lifespan of 24 and 13%, respectively. These beneficial effects were not observed with a catalytically inactive TERT, demonstrating that they require telomerase activity. Together, these results constitute a proof-of-principle of a role of TERT in delaying physiological aging and extending longevity in normal mice through a telomerase-based treatment, and demonstrate the feasibility of anti-aging gene therapy.
- ↑ Kouzarides T: Chromatin modifications and their function. Cell 2007. (PMID 17320507) [PubMed] [DOI] The surface of nucleosomes is studded with a multiplicity of modifications. At least eight different classes have been characterized to date and many different sites have been identified for each class. Operationally, modifications function either by disrupting chromatin contacts or by affecting the recruitment of nonhistone proteins to chromatin. Their presence on histones can dictate the higher-order chromatin structure in which DNA is packaged and can orchestrate the ordered recruitment of enzyme complexes to manipulate DNA. In this way, histone modifications have the potential to influence many fundamental biological processes, some of which may be epigenetically inherited.
- ↑ Mostoslavsky R et al.: Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006. (PMID 16439206) [PubMed] [DOI] The Sir2 histone deacetylase functions as a chromatin silencer to regulate recombination, genomic stability, and aging in budding yeast. Seven mammalian Sir2 homologs have been identified (SIRT1-SIRT7), and it has been speculated that some may have similar functions to Sir2. Here, we demonstrate that SIRT6 is a nuclear, chromatin-associated protein that promotes resistance to DNA damage and suppresses genomic instability in mouse cells, in association with a role in base excision repair (BER). SIRT6-deficient mice are small and at 2-3 weeks of age develop abnormalities that include profound lymphopenia, loss of subcutaneous fat, lordokyphosis, and severe metabolic defects, eventually dying at about 4 weeks. We conclude that one function of SIRT6 is to promote normal DNA repair, and that SIRT6 loss leads to abnormalities in mice that overlap with aging-associated degenerative processes.
- ↑ Herranz D et al.: Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 2010. (PMID 20975665) [PubMed] [DOI] [Full text] Genetic overexpression of protein deacetylase Sir2 increases longevity in a variety of lower organisms, and this has prompted interest in the effects of its closest mammalian homologue, Sirt1, on ageing and cancer. We have generated transgenic mice moderately overexpressing Sirt1 under its own regulatory elements (Sirt1-tg). Old Sirt1-tg mice present lower levels of DNA damage, decreased expression of the ageing-associated gene p16(Ink4a), a better general health and fewer spontaneous carcinomas and sarcomas. These effects, however, were not sufficiently potent to affect longevity. To further extend these observations, we developed a metabolic syndrome-associated liver cancer model in which wild-type mice develop multiple carcinomas. Sirt1-tg mice show a reduced susceptibility to liver cancer and exhibit improved hepatic protection from both DNA damage and metabolic damage. Together, these results provide direct proof of the anti-ageing activity of Sirt1 in mammals and of its tumour suppression activity in ageing- and metabolic syndrome-associated cancer.
- ↑ Brown K et al.: SIRT3 reverses aging-associated degeneration. Cell Rep 2013. (PMID 23375372) [PubMed] [DOI] [Full text] Despite recent controversy about their function in some organisms, sirtuins are thought to play evolutionarily conserved roles in lifespan extension. Whether sirtuins can reverse aging-associated degeneration is unknown. Tissue-specific stem cells persist throughout the entire lifespan to repair and maintain tissues, but their self-renewal and differentiation potential become dysregulated with aging. We show that SIRT3, a mammalian sirtuin that regulates the global acetylation landscape of mitochondrial proteins and reduces oxidative stress, is highly enriched in hematopoietic stem cells (HSCs) where it regulates a stress response. SIRT3 is dispensable for HSC maintenance and tissue homeostasis at a young age under homeostatic conditions but is essential under stress or at an old age. Importantly, SIRT3 is suppressed with aging, and SIRT3 upregulation in aged HSCs improves their regenerative capacity. Our study illuminates the plasticity of mitochondrial homeostasis controlling stem cell and tissue maintenance during the aging process and shows that aging-associated degeneration can be reversed by a sirtuin.
- ↑ Kanfi Y et al.: The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012. (PMID 22367546) [PubMed] [DOI] The significant increase in human lifespan during the past century confronts us with great medical challenges. To meet these challenges, the mechanisms that determine healthy ageing must be understood and controlled. Sirtuins are highly conserved deacetylases that have been shown to regulate lifespan in yeast, nematodes and fruitflies. However, the role of sirtuins in regulating worm and fly lifespan has recently become controversial. Moreover, the role of the seven mammalian sirtuins, SIRT1 to SIRT7 (homologues of the yeast sirtuin Sir2), in regulating lifespan is unclear. Here we show that male, but not female, transgenic mice overexpressing Sirt6 (ref. 4) have a significantly longer lifespan than wild-type mice. Gene expression analysis revealed significant differences between male Sirt6-transgenic mice and male wild-type mice: transgenic males displayed lower serum levels of insulin-like growth factor 1 (IGF1), higher levels of IGF-binding protein 1 and altered phosphorylation levels of major components of IGF1 signalling, a key pathway in the regulation of lifespan. This study shows the regulation of mammalian lifespan by a sirtuin family member and has important therapeutic implications for age-related diseases.
- ↑ 26.0 26.1 26.2 Larson K et al.: Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 2012. (PMID 22291607) [PubMed] [DOI] [Full text] Organismal aging is influenced by a multitude of intrinsic and extrinsic factors, and heterochromatin loss has been proposed to be one of the causes of aging. However, the role of heterochromatin in animal aging has been controversial. Here we show that heterochromatin formation prolongs lifespan and controls ribosomal RNA synthesis in Drosophila. Animals with decreased heterochromatin levels exhibit a dramatic shortening of lifespan, whereas increasing heterochromatin prolongs lifespan. The changes in lifespan are associated with changes in muscle integrity. Furthermore, we show that heterochromatin levels decrease with normal aging and that heterochromatin formation is essential for silencing rRNA transcription. Loss of epigenetic silencing and loss of stability of the rDNA locus have previously been implicated in aging of yeast. Taken together, these results suggest that epigenetic preservation of genome stability, especially at the rDNA locus, and repression of unnecessary rRNA synthesis, might be an evolutionarily conserved mechanism for prolonging lifespan.
- ↑ Wilson VL et al.: Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem 1987. (PMID 3611071) [PubMed] Significant losses of DNA 5-methyldeoxycytidine residues in old age could disrupt cellular gene expression and contribute to the physiological decline of the animal. Thus, the 5-methyldeoxycytidine content of DNAs, isolated from the tissues of two rodent species of various ages, were determined. Mus musculus lost DNA methylation sites at a rate of about 4.7 X 10(4) (approximately 0.012% of the newborn level)/month. Peromyscus leucopus lost DNA 5-methyldeoxycytidine residues at a rate of only 2.3 X 10(4) (approximately 0.006% of the newborn level)/month. Since P. leucopus generally live twice as long as M. musculus, the rate of loss of DNA 5-methyldeoxycytidine residues appears to be inversely related to life span. Similar losses in genomic 5-methyldeoxycytidine content were also observed to correlate with donor age in cultured normal human bronchial epithelial cells.
- ↑ 28.0 28.1 Heyn H et al.: Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A 2012. (PMID 22689993) [PubMed] [DOI] [Full text] Human aging cannot be fully understood in terms of the constrained genetic setting. Epigenetic drift is an alternative means of explaining age-associated alterations. To address this issue, we performed whole-genome bisulfite sequencing (WGBS) of newborn and centenarian genomes. The centenarian DNA had a lower DNA methylation content and a reduced correlation in the methylation status of neighboring cytosine--phosphate--guanine (CpGs) throughout the genome in comparison with the more homogeneously methylated newborn DNA. The more hypomethylated CpGs observed in the centenarian DNA compared with the neonate covered all genomic compartments, such as promoters, exonic, intronic, and intergenic regions. For regulatory regions, the most hypomethylated sequences in the centenarian DNA were present mainly at CpG-poor promoters and in tissue-specific genes, whereas a greater level of DNA methylation was observed in CpG island promoters. We extended the study to a larger cohort of newborn and nonagenarian samples using a 450,000 CpG-site DNA methylation microarray that reinforced the observation of more hypomethylated DNA sequences in the advanced age group. WGBS and 450,000 analyses of middle-age individuals demonstrated DNA methylomes in the crossroad between the newborn and the nonagenarian/centenarian groups. Our study constitutes a unique DNA methylation analysis of the extreme points of human life at a single-nucleotide resolution level.
- ↑ Zhang J et al.: Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation. Science 2022. (PMID 35143257) [PubMed] [DOI] Bivalent genes are ready for activation upon the arrival of developmental cues. Here, we report that BEND3 is a CpG island (CGI)-binding protein that is enriched at regulatory elements. The cocrystal structure of BEND3 in complex with its target DNA reveals the structural basis for its DNA methylation-sensitive binding property. Mouse embryos ablated of Bend3 died at the pregastrulation stage. Bend3 null embryonic stem cells (ESCs) exhibited severe defects in differentiation, during which hundreds of CGI-containing bivalent genes were prematurely activated. BEND3 is required for the stable association of polycomb repressive complex 2 (PRC2) at bivalent genes that are highly occupied by BEND3, which suggests a reining function of BEND3 in maintaining high levels of H3K27me3 at these bivalent genes in ESCs to prevent their premature activation in the forthcoming developmental stage.
- ↑ Seale K et al.: Making sense of the ageing methylome. Nat Rev Genet 2022. (PMID 35501397) [PubMed] [DOI] [Full text] Over time, the human DNA methylation landscape accrues substantial damage, which has been associated with a broad range of age-related diseases, including cardiovascular disease and cancer. Various age-related DNA methylation changes have been described, including at the level of individual CpGs, such as differential and variable methylation, and at the level of the whole methylome, including entropy and correlation networks. Here, we review these changes in the ageing methylome as well as the statistical tools that can be used to quantify them. We detail the evidence linking DNA methylation to ageing phenotypes and the longevity strategies aimed at altering both DNA methylation patterns and machinery to extend healthspan and lifespan. Lastly, we discuss theories on the mechanistic causes of epigenetic ageing.
- ↑ Osorio FG et al.: Nuclear envelope alterations generate an aging-like epigenetic pattern in mice deficient in Zmpste24 metalloprotease. Aging Cell 2010. (PMID 20961378) [PubMed] [DOI] Mutations in the nuclear envelope protein lamin A or in its processing protease ZMPSTE24 cause human accelerated aging syndromes, including Hutchinson-Gilford progeria syndrome. Similarly, Zmpste24-deficient mice accumulate unprocessed prelamin A and develop multiple progeroid symptoms, thus representing a valuable animal model for the study of these syndromes. Zmpste24-deficient mice also show marked transcriptional alterations associated with chromatin disorganization, but the molecular links between both processes are unknown. We report herein that Zmpste24-deficient mice show a hypermethylation of rDNA that reduces the transcription of ribosomal genes, being this reduction reversible upon treatment with DNA methyltransferase inhibitors. This alteration has been previously described during physiological aging in rodents, suggesting its potential role in the development of the progeroid phenotypes. We also show that Zmpste24-deficient mice present global hypoacetylation of histones H2B and H4. By using a combination of RNA sequencing and chromatin immunoprecipitation assays, we demonstrate that these histone modifications are associated with changes in the expression of several genes involved in the control of cell proliferation and metabolic processes, which may contribute to the plethora of progeroid symptoms exhibited by Zmpste24-deficient mice. The identification of these altered genes may help to clarify the molecular mechanisms underlying aging and progeroid syndromes as well as to define new targets for the treatment of these dramatic diseases.
- ↑ Shumaker DK et al.: Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A 2006. (PMID 16738054) [PubMed] [DOI] [Full text] The premature aging disease Hutchinson-Gilford Progeria Syndrome (HGPS) is caused by a mutant lamin A (LADelta50). Nuclei in cells expressing LADelta50 are abnormally shaped and display a loss of heterochromatin. To determine the mechanisms responsible for the loss of heterochromatin, epigenetic marks regulating either facultative or constitutive heterochromatin were examined. In cells from a female HGPS patient, histone H3 trimethylated on lysine 27 (H3K27me3), a mark for facultative heterochromatin, is lost on the inactive X chromosome (Xi). The methyltransferase responsible for this mark, EZH2, is also down-regulated. These alterations are detectable before the changes in nuclear shape that are considered to be the pathological hallmarks of HGPS cells. The results also show a down-regulation of the pericentric constitutive heterochromatin mark, histone H3 trimethylated on lysine 9, and an altered association of this mark with heterochromatin protein 1alpha (Hp1alpha) and the CREST antigen. This loss of constitutive heterochromatin is accompanied by an up-regulation of pericentric satellite III repeat transcripts. In contrast to these decreases in histone H3 methylation states, there is an increase in the trimethylation of histone H4K20, an epigenetic mark for constitutive heterochromatin. Expression of LADelta50 in normal cells induces changes in histone methylation patterns similar to those seen in HGPS cells. The epigenetic changes described most likely represent molecular mechanisms responsible for the rapid progression of premature aging in HGPS patients.
- ↑ Dang W et al.: Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 2009. (PMID 19516333) [PubMed] [DOI] [Full text] Cells undergoing developmental processes are characterized by persistent non-genetic alterations in chromatin, termed epigenetic changes, represented by distinct patterns of DNA methylation and histone post-translational modifications. Sirtuins, a group of conserved NAD(+)-dependent deacetylases or ADP-ribosyltransferases, promote longevity in diverse organisms; however, their molecular mechanisms in ageing regulation remain poorly understood. Yeast Sir2, the first member of the family to be found, establishes and maintains chromatin silencing by removing histone H4 lysine 16 acetylation and bringing in other silencing proteins. Here we report an age-associated decrease in Sir2 protein abundance accompanied by an increase in H4 lysine 16 acetylation and loss of histones at specific subtelomeric regions in replicatively old yeast cells, which results in compromised transcriptional silencing at these loci. Antagonizing activities of Sir2 and Sas2, a histone acetyltransferase, regulate the replicative lifespan through histone H4 lysine 16 at subtelomeric regions. This pathway, distinct from existing ageing models for yeast, may represent an evolutionarily conserved function of sirtuins in regulation of replicative ageing by maintenance of intact telomeric chromatin.
- ↑ Hu Z et al.: Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 2014. (PMID 24532716) [PubMed] [DOI] [Full text] All eukaryotic cells divide a finite number of times, although the mechanistic basis of this replicative aging remains unclear. Replicative aging is accompanied by a reduction in histone protein levels, and this is a cause of aging in budding yeast. Here we show that nucleosome occupancy decreased by 50% across the whole genome during replicative aging using spike-in controlled micrococcal nuclease digestion followed by sequencing. Furthermore, nucleosomes became less well positioned or moved to sequences predicted to better accommodate histone octamers. The loss of histones during aging led to transcriptional induction of all yeast genes. Genes that are normally repressed by promoter nucleosomes were most induced, accompanied by preferential nucleosome loss from their promoters. We also found elevated levels of DNA strand breaks, mitochondrial DNA transfer to the nuclear genome, large-scale chromosomal alterations, translocations, and retrotransposition during aging.
- ↑ O'Sullivan RJ et al.: Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 2010. (PMID 20890289) [PubMed] [DOI] [Full text] During replicative aging of primary cells morphological transformations occur, the expression pattern is altered and chromatin changes globally. Here we show that chronic damage signals, probably caused by telomere processing, affect expression of histones and lead to their depletion. We investigated the abundance and cell cycle expression of histones and histone chaperones and found defects in histone biosynthesis during replicative aging. Simultaneously, epigenetic marks were redistributed across the phases of the cell cycle and the DNA damage response (DDR) machinery was activated. The age-dependent reprogramming affected telomeric chromatin itself, which was progressively destabilized, leading to a boost of the telomere-associated DDR with each successive cell cycle. We propose a mechanism in which changes in the structural and epigenetic integrity of telomeres affect core histones and their chaperones, enforcing a self-perpetuating pathway of global epigenetic changes that ultimately leads to senescence.
- ↑ Bahar R et al.: Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 2006. (PMID 16791200) [PubMed] [DOI] The accumulation of somatic DNA damage has been implicated as a cause of ageing in metazoa. One possible mechanism by which increased DNA damage could lead to cellular degeneration and death is by stochastic deregulation of gene expression. Here we directly test for increased transcriptional noise in aged tissue by dissociating single cardiomyocytes from fresh heart samples of both young and old mice, followed by global mRNA amplification and quantification of mRNA levels in a panel of housekeeping and heart-specific genes. Although gene expression levels already varied among cardiomyocytes from young heart, this heterogeneity was significantly elevated at old age. We had demonstrated previously an increased load of genome rearrangements and other mutations in the heart of aged mice. To confirm that increased stochasticity of gene expression could be a result of increased genome damage, we treated mouse embryonic fibroblasts in culture with hydrogen peroxide. Such treatment resulted in a significant increase in cell-to-cell variation in gene expression, which was found to parallel the induction and persistence of genome rearrangement mutations at a lacZ reporter locus. These results underscore the stochastic nature of the ageing process, and could provide a mechanism for age-related cellular degeneration and death in tissues of multicellular organisms.
- ↑ Harries LW et al.: Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell 2011. (PMID 21668623) [PubMed] [DOI] [Full text] Aging is a major risk factor for chronic disease in the human population, but there are little human data on gene expression alterations that accompany the process. We examined human peripheral blood leukocyte in-vivo RNA in a large-scale transcriptomic microarray study (subjects aged 30-104 years). We tested associations between probe expression intensity and advancing age (adjusting for confounding factors), initially in a discovery set (n= 58), following-up findings in a replication set (n=240). We confirmed expression of key results by real-time PCR. Of 16,571 expressed probes, only 295 (2%) were robustly associated with age. Just six probes were required for a highly efficient model for distinguishing between young and old (area under the curve in replication set; 95%). The focused nature of age-related gene expression may therefore provide potential biomarkers of aging. Similarly, only 7 of 1065 biological or metabolic pathways were age-associated, in gene set enrichment analysis, notably including the processing of messenger RNAs (mRNAs); [P<0.002, false discovery rate (FDR) q<0.05]. This is supported by our observation of age-associated disruption to the balance of alternatively expressed isoforms for selected genes, suggesting that modification of mRNA processing may be a feature of human aging.
- ↑ Nicholas A et al.: Age-related gene-specific changes of A-to-I mRNA editing in the human brain. Mech Ageing Dev 2010. (PMID 20538013) [PubMed] [DOI] [Full text] A-to-I editing is an adenosine-to-inosine modification of mRNA particularly widespread in the human brain, where it affects thousands of genes. A growing body of evidence suggests that A-to-I RNA editing is necessary for normal development and maintenance in mammals and that its deficiencies contribute to a number of pathological states. In this study, we examined whether mRNA editing levels of two mRNA species, CYFIP2 and GABRA3, change with aging. CYFIP2 has been implicated in synaptic maintenance, while GABRA3 is a GABA receptor subunit, a part of the major inhibitory neurotransmitter system in the CNS. The levels of mRNA editing were assessed in cortex samples of 20 subjects 22-102 years old. The data show an age-dependent statistically significant decrease in editing in CYFIP2. GABRA3 editing remained much more stable with age, implying that age-related decline of RNA editing is gene-specific. This is the first report of age-dependent decline in A-to-I editing. Further examination of these and other vulnerable genes may reveal specific RNA editing mechanisms that contribute to the aging phenotype.
- ↑ Toledano H et al.: The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 2012. (PMID 22660319) [PubMed] [DOI] [Full text] Adult stem cells support tissue homeostasis and repair throughout the life of an individual. During ageing, numerous intrinsic and extrinsic changes occur that result in altered stem-cell behaviour and reduced tissue maintenance and regeneration. In the Drosophila testis, ageing results in a marked decrease in the self-renewal factor Unpaired (Upd), leading to a concomitant loss of germline stem cells. Here we demonstrate that IGF-II messenger RNA binding protein (Imp) counteracts endogenous small interfering RNAs to stabilize upd (also known as os) RNA. However, similar to upd, Imp expression decreases in the hub cells of older males, which is due to the targeting of Imp by the heterochronic microRNA let-7. In the absence of Imp, upd mRNA therefore becomes unprotected and susceptible to degradation. Understanding the mechanistic basis for ageing-related changes in stem-cell behaviour will lead to the development of strategies to treat age-onset diseases and facilitate stem-cell-based therapies in older individuals.
- ↑ Ugalde AP et al.: Micromanaging aging with miRNAs: new messages from the nuclear envelope. Nucleus 2011. (PMID 22064465) [PubMed] [DOI] Over the last years, the discovery of microRNAs (miRNAs) has revolutionized the classic concepts of gene expression regulation and has introduced a new group of molecules that may contribute to the complex changes observed during aging. Although several Caenorhabditis elegans miRNAs have been proved to influence the nematode life span, the current knowledge about miRNA-mediated regulation of mammalian aging is still limited. Recently, we have analyzed the functional relevance of miRNAs in accelerate aging by using Zmpste24-/- mice, a murine model that phenocopies Hutchinson-Gilford progeria syndrome. These studies have revealed that the nuclear abnormalities present in these mice affect the expression levels of several miRNAs, including a marked upregulation of miR-1 and miR-29. Furthermore, we have found that the altered expression of these miRNAs may contribute to the progeroid phenotype of mutant mice by modulating the levels of key components of the somatroph axis and DNA damage response pathways. Here, we discuss these recent discoveries and summarize the present evidences regarding the involvement of aging-associated miRNAs or geromiRs in senescence and longevity regulation.
- ↑ 41.0 41.1 Liu N et al.: The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 2012. (PMID 22343898) [PubMed] [DOI] [Full text] Human neurodegenerative diseases have the temporal hallmark of afflicting the elderly population. Ageing is one of the most prominent factors to influence disease onset and progression, yet little is known about the molecular pathways that connect these processes. To understand this connection it is necessary to identify the pathways that functionally integrate ageing, chronic maintenance of the brain and modulation of neurodegenerative disease. MicroRNAs (miRNA) are emerging as critical factors in gene regulation during development; however, their role in adult-onset, age-associated processes is only beginning to be revealed. Here we report that the conserved miRNA miR-34 regulates age-associated events and long-term brain integrity in Drosophila, providing a molecular link between ageing and neurodegeneration. Fly mir-34 expression exhibits adult-onset, brain-enriched and age-modulated characteristics. Whereas mir-34 loss triggers a gene profile of accelerated brain ageing, late-onset brain degeneration and a catastrophic decline in survival, mir-34 upregulation extends median lifespan and mitigates neurodegeneration induced by human pathogenic polyglutamine disease protein. Some of the age-associated effects of miR-34 require adult-onset translational repression of Eip74EF, an essential ETS domain transcription factor involved in steroid hormone pathways. Our studies indicate that miRNA-dependent pathways may have an impact on adult-onset, age-associated events by silencing developmental genes that later have a deleterious influence on adult life cycle and disease, and highlight fly miR-34 as a key miRNA with a role in this process.
- ↑ 42.0 42.1 Shen Y et al.: A steroid receptor-microRNA switch regulates life span in response to signals from the gonad. Science 2012. (PMID 23239738) [PubMed] [DOI] [Full text] Although the gonad primarily functions in procreation, it also affects animal life span. Here, we show that removal of the Caenorhabditis elegans germ line triggers a switch in the regulatory state of the organism to promote longevity, co-opting components involved in larval developmental timing circuits. These components include the DAF-12 steroid receptor, which is involved in the larval stage two-to-stage three (L2-L3) transition and up-regulates members of the let-7 microRNA (miRNA) family. The miRNAs target an early larval nuclear factor lin-14 and akt-1/kinase, thereby stimulating DAF-16/FOXO signaling to extend life. Our studies suggest that metazoan life span is coupled to the gonad through elements of a developmental timer.
- ↑ 43.0 43.1 Smith-Vikos T & Slack FJ: MicroRNAs and their roles in aging. J Cell Sci 2012. (PMID 22294612) [PubMed] [DOI] [Full text] MicroRNAs (miRNAs) are a class of short non-coding RNAs that bind mRNAs through partial base-pair complementarity with their target genes, resulting in post-transcriptional repression of gene expression. The role of miRNAs in controlling aging processes has been uncovered recently with the discovery of miRNAs that regulate lifespan in the nematode Caenorhabditis elegans through insulin and insulin-like growth factor-1 signaling and DNA damage checkpoint factors. Furthermore, numerous miRNAs are differentially expressed during aging in C. elegans, but the specific functions of many of these miRNAs are still unknown. Recently, various miRNAs have been identified that are up- or down-regulated during mammalian aging by comparing their tissue-specific expression in younger and older mice. In addition, many miRNAs have been implicated in governing senescence in a variety of human cell lines, and the precise functions of some of these miRNAs in regulating cellular senescence have helped to elucidate mechanisms underlying aging. In this Commentary, we review the various regulatory roles of miRNAs during aging processes. We highlight how certain miRNAs can regulate aging on the level of organism lifespan, tissue aging or cellular senescence. Finally, we discuss future approaches that might be used to investigate the mechanisms by which miRNAs govern aging processes.
- ↑ 44.0 44.1 Powers ET et al.: Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 2009. (PMID 19298183) [PubMed] [DOI] Many diseases appear to be caused by the misregulation of protein maintenance. Such diseases of protein homeostasis, or "proteostasis," include loss-of-function diseases (cystic fibrosis) and gain-of-toxic-function diseases (Alzheimer's, Parkinson's, and Huntington's disease). Proteostasis is maintained by the proteostasis network, which comprises pathways that control protein synthesis, folding, trafficking, aggregation, disaggregation, and degradation. The decreased ability of the proteostasis network to cope with inherited misfolding-prone proteins, aging, and/or metabolic/environmental stress appears to trigger or exacerbate proteostasis diseases. Herein, we review recent evidence supporting the principle that proteostasis is influenced both by an adjustable proteostasis network capacity and protein folding energetics, which together determine the balance between folding efficiency, misfolding, protein degradation, and aggregation. We review how small molecules can enhance proteostasis by binding to and stabilizing specific proteins (pharmacologic chaperones) or by increasing the proteostasis network capacity (proteostasis regulators). We propose that such therapeutic strategies, including combination therapies, represent a new approach for treating a range of diverse human maladies.
- ↑ Min JN et al.: CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol 2008. (PMID 18411298) [PubMed] [DOI] [Full text] During the course of biological aging, there is a gradual accumulation of damaged proteins and a concomitant functional decline in the protein degradation system. Protein quality control is normally ensured by the coordinated actions of molecular chaperones and the protein degradation system that collectively help to maintain protein homeostasis. The carboxyl terminus of Hsp70-interacting protein (CHIP), a ubiquitin ligase/cochaperone, participates in protein quality control by targeting a broad range of chaperone substrates for proteasome degradation via the ubiquitin-proteasome system, demonstrating a broad involvement of CHIP in maintaining cytoplasmic protein quality control. In the present study, we have investigated the influence that protein quality control exerts on the aging process by using CHIP-/- mice. CHIP deficiency in mice leads to a markedly reduced life span, along with accelerated age-related pathophysiological phenotypes. These features were accompanied by indications of accelerated cellular senescence and increased indices of oxidative stress. In addition, CHIP-/- mice exhibit a deregulation of protein quality control, as indicated by elevated levels of toxic oligomer proteins and a decline in proteasome activity. Taken together, these data reveal that impaired protein quality control contributes to cellular senescence and implicates CHIP-dependent quality control mechanisms in the regulation of mammalian longevity in vivo.
- ↑ Morrow G et al.: Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J 2004. (PMID 14734639) [PubMed] [DOI] Heat shock proteins (Hsp) are involved in protein folding, transport and stress resistance. Studies reporting an increased mRNA level of hsp genes in aged Drosophila suggest that expression of Hsp might be beneficial in preventing damages induced by aging. Because oxidative damage is often observed in aged organisms and mitochondria are sensitive to reactive oxygen species, we tested the hypothesis that increased levels of a small Hsp localized in mitochondria, Hsp22 of Drosophila melanogaster, could protect mitochondrial proteins and influence the aging process. We demonstrate that a ubiquitous or a targeted expression of Hsp22 within motorneurons increases the mean life span by more than 30%. Hsp22 shows beneficial effects on early-aging events since the premortality phase displays the same increase as the mean lifespan. Moreover, flies expressing Hsp22 in their motorneurons maintain their locomotor activity longer as assessed by a negative geotaxis assay. The motorneurons-targeted expression of Hsp22 also significantly increases flies' resistance to oxidative injuries induced by paraquat (up to 35%) and thermal stress (39% at 30 degrees C and 23% at 37 degrees C). These observations establish Hsp22 as a key player in cell-protection mechanisms against oxidative injuries and aging in Drosophila and corroborate the pivotal role of mitochondria in the process of aging.
- ↑ Walker GA & Lithgow GJ: Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2003. (PMID 12882326) [PubMed] [DOI] Insulin-like signalling is a key determinate of lifespan in diverse species including mammals but the mechanism by which this pathway influences the rate of aging is unknown. In the roundworm Caenorhabditis elegans, mutations in the insulin-like signalling pathway extend adult lifespan and are associated with up-regulation of stress response genes including those for heat shock proteins (HSPs). We tested the hypothesis that the C. elegans insulin-like signalling pathway determines longevity through modulating HSP levels. We introduced extra copies of the gene encoding HSP-16 and this conferred stress resistance and longevity both in a wildtype and a long-lived mutant strain. The DAF-16 transcription factor is essential for maximal hsp-16 expression and for lifespan extension conferred by hsp-16. This demonstrates that lifespan is determined in part by insulin-like regulation of molecular chaperones.
- ↑ Kaushik S & Cuervo AM: The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 2018. (PMID 29626215) [PubMed] [DOI] [Full text] Chaperone-mediated autophagy (CMA) was the first studied process that indicated that degradation of intracellular components by the lysosome can be selective - a concept that is now well accepted for other forms of autophagy. Lysosomes can degrade cellular cytosol in a nonspecific manner but can also discriminate what to target for degradation with the involvement of a degradation tag, a chaperone and a sophisticated mechanism to make the selected proteins cross the lysosomal membrane through a dedicated translocation complex. Recent studies modulating CMA activity in vivo using transgenic mouse models have demonstrated that selectivity confers on CMA the ability to participate in the regulation of multiple cellular functions. Timely degradation of specific cellular proteins by CMA modulates, for example, glucose and lipid metabolism, DNA repair, cellular reprograming and the cellular response to stress. These findings expand the physiological relevance of CMA beyond its originally identified role in protein quality control and reveal that CMA failure with age may aggravate diseases, such as ageing-associated neurodegeneration and cancer.
- ↑ Wong SQ et al.: Autophagy in aging and longevity. Hum Genet 2020. (PMID 31144030) [PubMed] [DOI] [Full text] Our understanding of the process of autophagy and its role in health and diseases has grown remarkably in the last two decades. Early work established autophagy as a general bulk recycling process which involves the sequestration and transport of intracellular material to the lysosome for degradation. Currently, autophagy is viewed as a nexus of metabolic and proteostatic signalling that can determine key physiological decisions from cell fate to organismal lifespan. Here, we review the latest literature on the role of autophagy and lysosomes in stress response and longevity. We highlight the connections between autophagy and metabolic processes, the network associated with its regulation, and the links between autophagic dysfunction, neurodegenerative diseases, and aging.
- ↑ Aman Y et al.: Autophagy in healthy aging and disease. Nat Aging 2021. (PMID 34901876) [PubMed] [DOI] [Full text] Autophagy is a fundamental cellular process that eliminates molecules and subcellular elements, including nucleic acids, proteins, lipids and organelles, via lysosome-mediated degradation to promote homeostasis, differentiation, development and survival. While autophagy is intimately linked to health, the intricate relationship among autophagy, aging and disease remains unclear. This Review examines several emerging features of autophagy and postulates how they may be linked to aging as well as to the development and progression of disease. In addition, we discuss current preclinical evidence arguing for the use of autophagy modulators as suppressors of age-related pathologies such as neurodegenerative diseases. Finally, we highlight key questions and propose novel research avenues that will likely reveal new links between autophagy and the hallmarks of aging. Understanding the precise interplay between autophagy and the risk of age-related pathologies across organisms will eventually facilitate the development of clinical applications that promote long-term health.
- ↑ Fernández ÁF et al.: Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018. (PMID 29849149) [PubMed] [DOI] [Full text] Autophagy increases the lifespan of model organisms; however, its role in promoting mammalian longevity is less well-established1,2. Here we report lifespan and healthspan extension in a mouse model with increased basal autophagy. To determine the effects of constitutively increased autophagy on mammalian health, we generated targeted mutant mice with a Phe121Ala mutation in beclin 1 (Becn1F121A/F121A) that decreases its interaction with the negative regulator BCL2. We demonstrate that the interaction between beclin 1 and BCL2 is disrupted in several tissues in Becn1 F121A/F121A knock-in mice in association with higher levels of basal autophagic flux. Compared to wild-type littermates, the lifespan of both male and female knock-in mice is significantly increased. The healthspan of the knock-in mice also improves, as phenotypes such as age-related renal and cardiac pathological changes and spontaneous tumorigenesis are diminished. Moreover, mice deficient in the anti-ageing protein klotho 3 have increased beclin 1 and BCL2 interaction and decreased autophagy. These phenotypes, along with premature lethality and infertility, are rescued by the beclin 1(F121A) mutation. Together, our data demonstrate that disruption of the beclin 1-BCL2 complex is an effective mechanism to increase autophagy, prevent premature ageing, improve healthspan and promote longevity in mammals.
- ↑ Alsaleh G et al.: Autophagy in T cells from aged donors is maintained by spermidine and correlates with function and vaccine responses. Elife 2020. (PMID 33317695) [PubMed] [DOI] [Full text] Vaccines are powerful tools to develop immune memory to infectious diseases and prevent excess mortality. In older adults, however vaccines are generally less efficacious and the molecular mechanisms that underpin this remain largely unknown. Autophagy, a process known to prevent aging, is critical for the maintenance of immune memory in mice. Here, we show that autophagy is specifically induced in vaccine-induced antigen-specific CD8+ T cells in healthy human volunteers. In addition, reduced IFNγ secretion by RSV-induced T cells in older vaccinees correlates with low autophagy levels. We demonstrate that levels of the endogenous autophagy-inducing metabolite spermidine fall in human T cells with age. Spermidine supplementation in T cells from old donors recovers their autophagy level and function, similar to young donors' cells, in which spermidine biosynthesis has been inhibited. Finally, our data show that endogenous spermidine maintains autophagy via the translation factor eIF5A and transcription factor TFEB. In summary, we have provided evidence for the importance of autophagy in vaccine immunogenicity in older humans and uncovered two novel drug targets that may increase vaccination efficiency in the aging context.
- ↑ Researchers Propose Five New Hallmarks of Aging, https://www.lifespan.io/news/researchers-propose-five-new-hallmarks-of-aging/
- ↑ López-Otín C et al.: Meta-hallmarks of aging and cancer. Cell Metab 2023. (PMID 36599298) [PubMed] [DOI] Both aging and cancer are characterized by a series of partially overlapping "hallmarks" that we subject here to a meta-analysis. Several hallmarks of aging (i.e., genomic instability, epigenetic alterations, chronic inflammation, and dysbiosis) are very similar to specific cancer hallmarks and hence constitute common "meta-hallmarks," while other features of aging (i.e., telomere attrition and stem cell exhaustion) act likely to suppress oncogenesis and hence can be viewed as preponderantly "antagonistic hallmarks." Disabled macroautophagy and cellular senescence are two hallmarks of aging that exert context-dependent oncosuppressive and pro-tumorigenic effects. Similarly, the equivalence or antagonism between aging-associated deregulated nutrient-sensing and cancer-relevant alterations of cellular metabolism is complex. The agonistic and antagonistic relationship between the processes that drive aging and cancer has bearings for the age-related increase and oldest age-related decrease of cancer morbidity and mortality, as well as for the therapeutic management of malignant disease in the elderly.
